Fluorescence is great for showing the intricate structures of cells in spectacular colors. Yet, there are some situations where getting the necessary information without fluorescent labels is faster, less damaging—or just easier.
Deep-learning software can do this for you. Its convolutional neural networks can automatically recognize cells and nuclei from brightfield images. It can also train itself to improve its capabilities over time.
We call this self-learning microscopy, and we’ve integrated this concept into the latest version of our cellSens life science image analysis software. Here’s an example of how self-learning microscopy can help you.
From DAPI to Deep Learning: An Experiment
An established technique to perform segmentation and counting of cell nuclei is to label the nuclei with DAPI and analyze the fluorescence images (Figure 1, right). We ran an experiment to see if we can get the same result when we analyze unstained brightfield images (Figure 1, left) with our cellSens deep learning module.
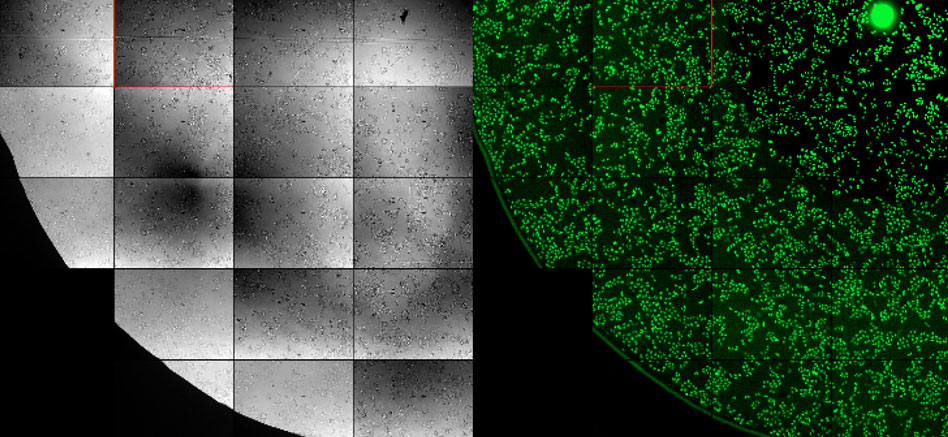
Figure 1: Brightfield image (left) and fluorescent image (right) of cell nuclei.
To train the software, we provided fluorescence and brightfield images of 40 positions in a 96-well plate. Thanks to self-learning microscopy, training was fully automatic and only took around 90 minutes.
The Results
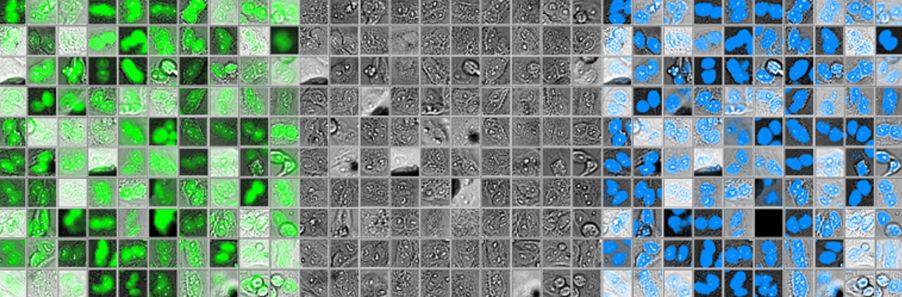
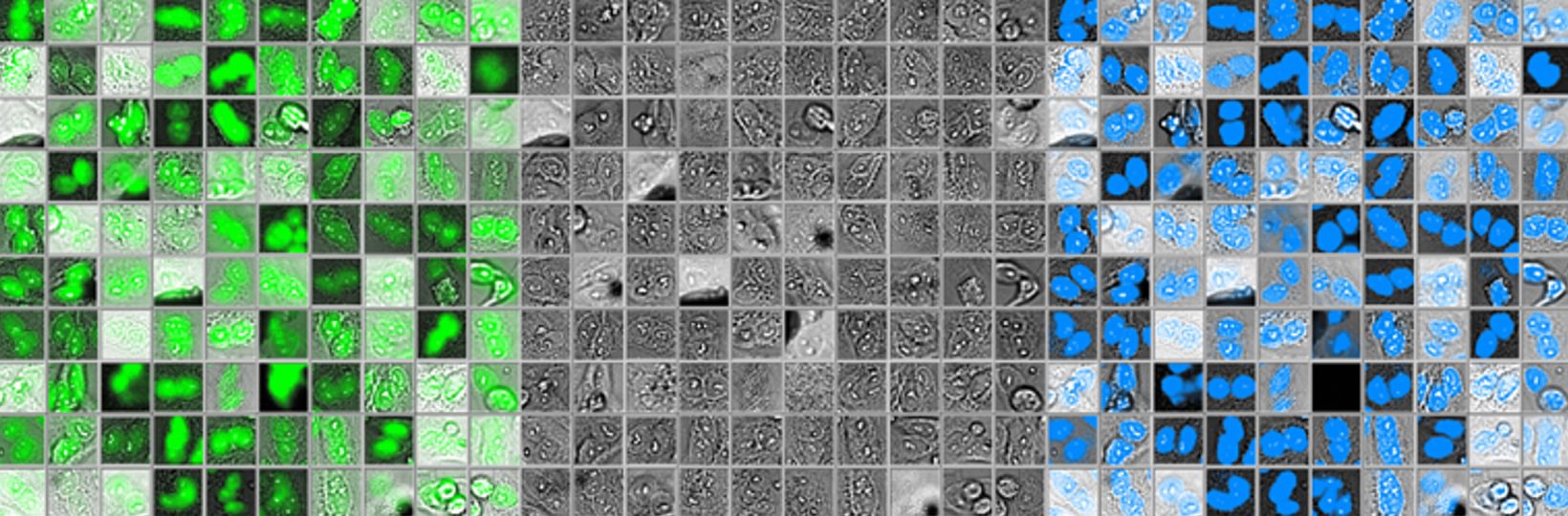
Figure 2 below shows 100 randomly selected nuclei detected using fluorescence (left), as well as the corresponding brightfield image (middle) and object shape predicted by deep learning (right). Although it shows excellent correspondence between the two methods, the total number of objects detected with deep learning was around 3% higher.
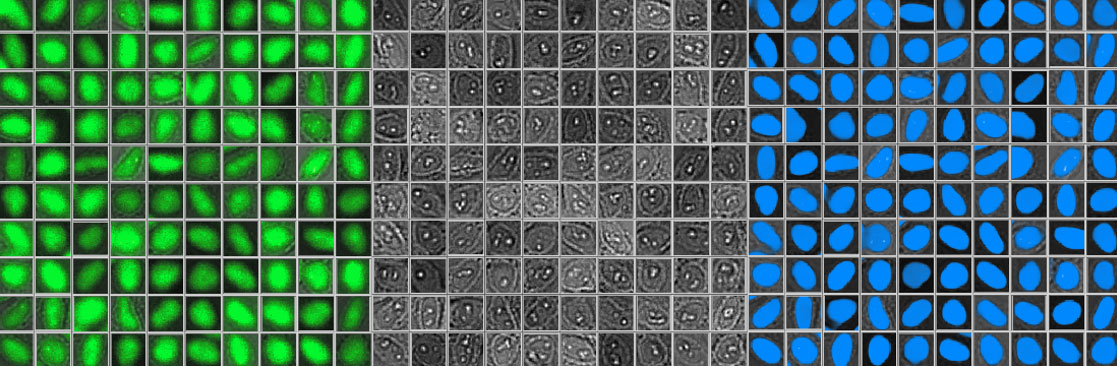
Figure 2: Nuclei detected using fluorescence (left), the corresponding brightfield image (middle), and object shape predicted by deep learning technology (right).
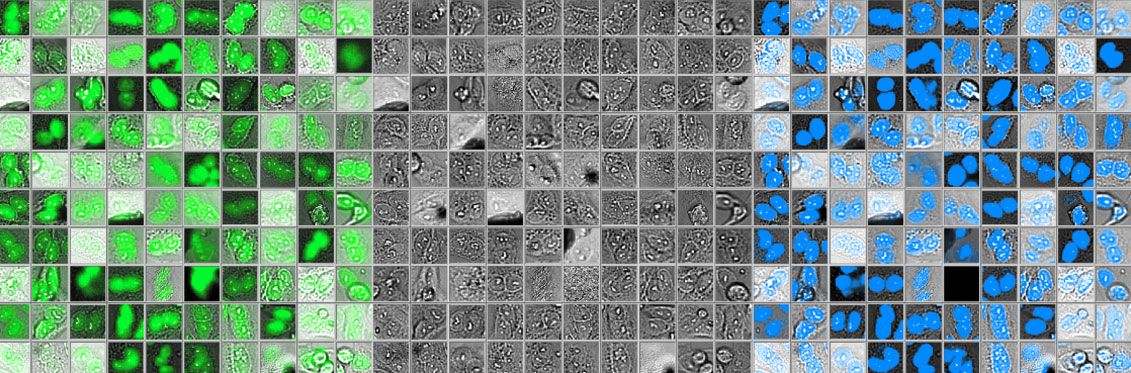
While investigating this difference, we found that the fluorescence data contained many unusually large objects. As we compared the images in this subset, we discovered that many of the fluorescent “objects” were actually two nuclei counted as one object—and most of them were correctly reported by the deep-learning software as two separate objects (see Figure 3 below).
Figure 3: Compared to the fluorescence image (left), the deep-learning software clearly distinguishes close nuclei from one another (right).
Important Lessons from this Self-Learning Microscopy Experiment
This experiment shows that image analysis methods based on deep-learning technology can easily rival or even perform better than methods based on fluorescence. And what’s more, the neural network training for accurate object detection is quick and automated.
The benefits to live cell imaging are clear: aside from the improved accuracy, image analysis powered by deep learning also means you don’t need genetic modifications (GFP) or nucleus markers. The result: you can save time on sample preparation, save a fluorescence channel for other markers, and reduce phototoxicity with shorter exposure times—what’s not to like?
Related Content
Video: The Power of Deep Learning for Self-Learning Microscopy
Brochure: cellSens Imaging Software for Life Science
5 Artificial Intelligence Lessons from a Year of Life Science Research