Accelerating and Optimizing the Segmentation and Analysis of Pancreatic Islets with the VS200 Research Slide Scanner and TruAI Deep-Learning Solution
To achieve relevant data for their investigations, researchers need to analyze large image sets. This analysis can be both quantitative and qualitative, and it must also be reliable and unbiased. Depending on the morphological area of interest, this task can be quite cumbersome. To accelerate this process, Olympus offers a deep-learning solution in combination with the SLIDEVIEW VS200 research slide scanner. To test this solution, we applied it to the segmentation and analysis of pancreatic islets.
Pancreatic Islet Detection and Segmentation
Working in experimental diabetes research, researchers aim to improve their understanding of the complex process of this disease to be able to develop better, more efficient treatments. An essential part of their research involves analyzing a significant number of pancreatic sections.
The pancreas has two major functions, excreting enzymes to break down the proteins, lipids, carbohydrates, and nucleic acids in food (exocrine) and secreting the hormones insulin and glucagon to control blood sugar levels (endocrine). The latter is relevant for diabetes. Groups of beta cells in the pancreas, also called islets of Langerhans, play a fundamental role in insulin secretion. Insulin helps lower glucose in your blood by stimulating its absorption into the cells of other tissues for energy.
In the investigation of diabetes and other metabolic diseases, researchers want to analyze the percentage and relative position of these insulin-containing beta cells. Microscopic analysis of a high volume of images is crucial to determine the morphology and to quantify the pancreatic islets of Langerhans.
Experiment Using the SLIDEVIEW VS200 Research Slide Scanner and TruAI Deep-Learning Solution
To demonstrate how Olympus TruAI deep-learning technology can improve this research application, we performed a project where we investigated the aging processes in islets of Langerhans in CB57BL/6NTac mice to understand the changes in islet morphology and function.
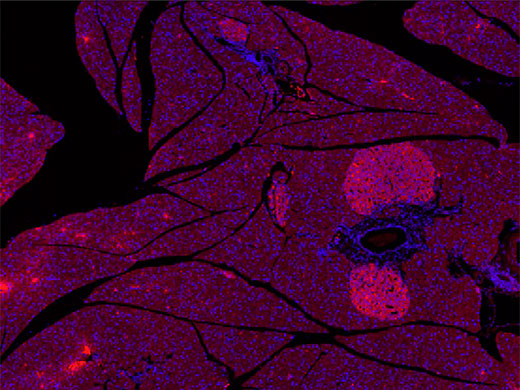
Example of a slide scanned by an Olympus VS200 slide scanner for our experiment. Pancreas section stained with Alexa 594 and DAPI Tissue sections are courtesy of Univ.-Prof. Dr. rer. nat.Simone E. Baltrusch from the Institute for Medical Biochemistry and Molecular Biology at Rostock University Medical Center in Rostock, Germany
The samples, courtesy of Professor Simone E. Baltrusch and Dr. Cindy Zehm from the Institute for Medical Biochemistry and Molecular Biology at Rostock University Medical Center, Germany, are mouse pancreatic sections prepared with an insulin antibody staining protocol. This staining enables beta cells within the islets of Langerhans to be identified. These cells correspond to the endocrine part of the pancreas, which differs in the cell architecture from the exocrine part.
Digital slides of the mouse pancreas samples were acquired rapidly using an Olympus SLIDEVIEW VS200 research slide scanner at 10× magnification. The result was 40 images in which the islets of Langerhans could be easily identified with the human eye. The images clearly showed the red, Alexa 594-stained secondary antibody attached to the primary antibody for insulin-producing beta cells, contrasting against the DAPI (blue) nuclei counterstain.
Challenges of Automatically Detecting and Analyzing Pancreatic Islets
Visualizing pancreatic islets in a murine pancreatic section sample can be done using specific fluorescence antibodies. To analyze these structures in scanned images, normally researchers select the islets manually, which is highly time consuming. Moreover, traditional automatic segmentation methods, such as threshold-based algorithms, fail to exclusively detect pancreatic islets, as shown below.
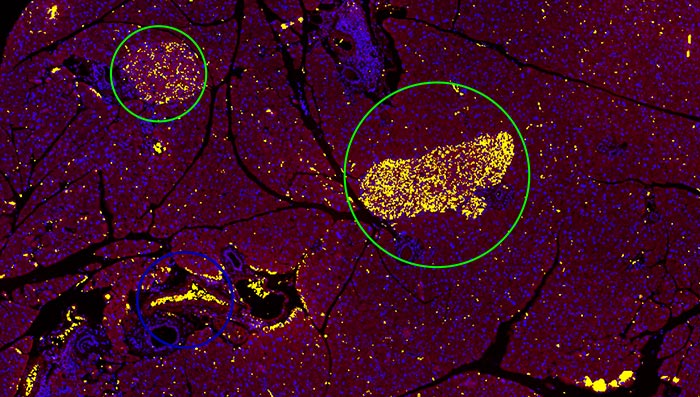
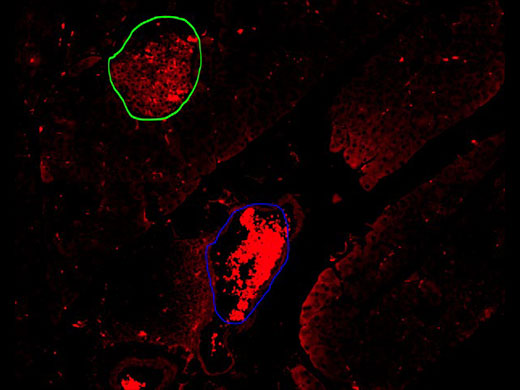
Image at 10x showing the detection based on the conventional threshold method in yellow, failing to distinguish the islets (green circle) from red blood cells (blue circle).
While observing the Alexa 594 channel, the autofluorescence of the erythrocytes was also detected, making it difficult to automatically distinguish the marked antibody used to stain the beta cells on a pancreatic islet (green circle) from a blood vessel full of erythrocytes (blue circle).The VS200 research slide scanner offers a TruAI module, a deep-learning approach based on convolutional neural networks, as an add-on to the VS-Desktop software for image analysis. These networks are a form of self-learning microscopy and are an extremely powerful technology for object segmentation. Thanks to this technology, we can automate the detection of the murine pancreatic islets. The neural network training process is explained in the next section. |
Pancreatic islet (green circle); blood cells (blue circle) |
Easy Workflow to Automatically Segment and Analyze Samples Using the TruAI Deep-Learning Solution
The first step toward this automatic analysis is to provide the software with annotated sample images or “ground truth” data. This was created by hand labeling the islets of twelve different mouse pancreas samples (green circle).
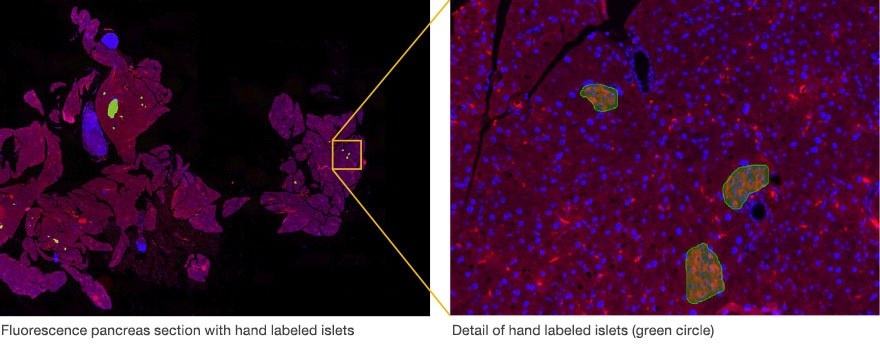
The more objects that are marked, the better. Higher numbers of objects with differences in intensity, color, size, and shape make the neural network more robust. The easy-to-use software add-on has various tools to facilitate the hand labeling.
After the labeled data is generated, the next step is training the deep neural network (DNN). In this phase, the network compares the ground truth data with its own calculated data until a high probability value is reached. This calculated data is a type of artificial intelligence (AI) that imitates the human brain (the so-called DNN), which learns to recognize structures and make intelligent judgements.
Lastly, the calculated DNN is applied, in this case, to the rest of the pancreas images to automatically detect and separate the islets of Langerhans.
A trained Olympus TruAI deep neural network can be transferred and used at any VS-Desktop station as well as at other compatible Olympus products.
This automated process for segmenting and analyzing pancreatic islets includes these three simple steps:
1. A new image is scanned with the SLIDEVIEW VS200 research slide scanner.
|
|
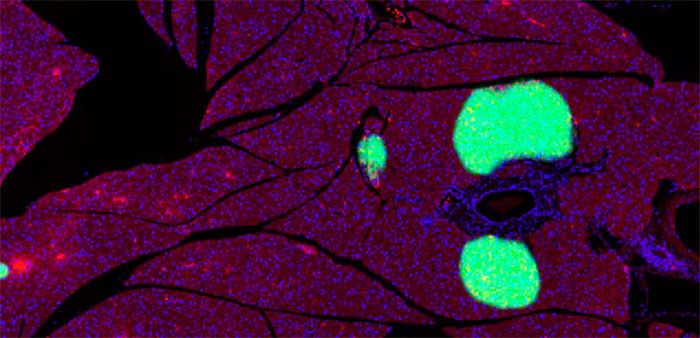
2. Pancreatic islets are detected and separated by the trained DNN.
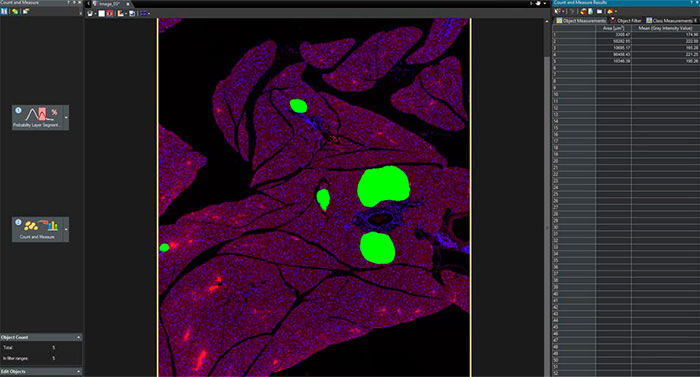
3. The detected pancreatic islets can be segmented and used to perform further Count and Measure analysis on the output image.
Summary of the Advantages of the SLIDEVIEW VS200 Research Slide Scanner with TruAI Deep-Learning Solution for the Detection and Segmentation of Pancreatic Islets
The TruAI module enabled easy object detection and segmentation of pancreatic islets from complex images with much higher reliability and accuracy than other existing automatic methods. Further analysis, such as counting and measurement, can also be performed based on these segmentation results.
The combination of the SLIDEVIEW VS200 research slide scanner and TruAI deep-learning solution can provide a complete workflow from the sample acquisition to the precise quantitative data analysis in a wide range of biological applications on a variety of images, such as cells and tissue samples in brightfield and fluorescence.
Precise automation of image analysis will relieve a lot of the tedious manual work for scientists, making their research more efficient.
Acknowledgements
This application note was prepared with the help of researchers from the Institute for Medical Biochemistry and Molecular Biology, University Medicine, University of Rostock, Rostock, Germany and Sara Quinones Gonzalez, Product Manager, Olympus Soft Imaging Solutions in Münster, Germany:
- Univ.-Prof. Dr. rer. nat. Simone E. Baltrusch, Institute for Medical Biochemistry and Molecular Biology, University Medicine, University of Rostock, Germany
- Dr. Cindy Zehm, Institute for Medical Biochemistry and Molecular Biology, University Medicine, University of Rostock, Germany
- Sara Quinones Gonzalez, Product Manager, Olympus Soft Imaging Solutions GmbH, Münster, Germany
Products Related to This Application
was successfully added to your bookmarks
Maximum Compare Limit of 5 Items
Please adjust your selection to be no more than 5 items to compare at once
Not Available in Your Country
Sorry, this page is not
available in your country.