3D Observation of Cleared Mouse Liver Using the FLUOVIEW FV3000 Microscope
Three-Dimensional Observation of Liver with High Resolution
3D observation of thick tissue specimens usually requires a two-photon excitation microscope due to the increased absorption and scattering of light that occurs when imaging deeper into the tissue sample. While the liver is typically a highly-scattering tissue, the use of clearing methods combined with the appropriate optics can enable thick tissue 3D observation using the FV3000 confocal microscope. In this experiment, the Olympus 30x silicone oil immersion objective with a numerical aperture (NA) of 1.05 and a working distance (WD) of 0.8 mm enabled high resolution 3D observation of the biliary tree structure in cleared mouse liver samples.
Experiment Summary
Optical Clearing of Mice Liver Tissue Specimens
Liver tissue collected from a mouse |
|
The tissue is stained for fluorescence by immunostaining |
|
The liver tissue is cleared using SeeDB |
Courtesy of: K. Kamimoto, K. Kaneko, CY. Kok, H. Okada, A. Miyajima, and T. Itoh, "Heterogeneity and stochastic growth regulation of biliary epithelial cells dictate dynamic epithelial tissue remodeling," Elife, 2016 Jul. 19;5, pii: e15034, doi: 10.7554/eLife.15034.
Experiment protocol for visualizing a complex biliary network in 3D. Following tissue collection, biliary tissues undergo immunostaining to visualize a complex biliary network in three dimensions. The immunostained specimens are then cleared using SeeDB1).
1) Reference:Ke MT, S. Fujimoto, and T. Imai, "SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction," Nat Neurosci, 2013 Aug; 16 (8): 1154–61, doi: 10.1038/nn.3447, Epub 2013 Jun. 23, PMID: 23792946
Three-Dimensional Observation of Biliary Tree Structures in Injured Mouse Liver with a 20x Objective (UPLSAPO20X (NA: 0.75, WD: 0.6 mm))
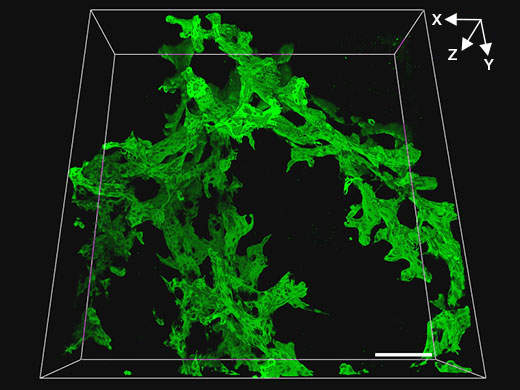 | The goal of the experiment was to quantitatively measure the length and thickness of the branches of biliary tree structures in injured mouse livers. For initial screening of samples, a 20x dry objective was used on the FV3000 to obtain consecutive, wide field-of-view tomographic images of biliary tissue (green, biliary epithelial cell marker CK19) in 200 µm thick cleared liver tissue specimens. This setup enabled effective quantitative observation for many samples, and enabled us to quickly find samples to examine at higher resolution. In the image above, the scale bar represents 100 µm. |
Three-Dimensional Observation of Biliary Tree Structures in Mouse Liver with a 30x Objective (UPLSAO30XS (NA: 1.05, WD: 0.8 mm))
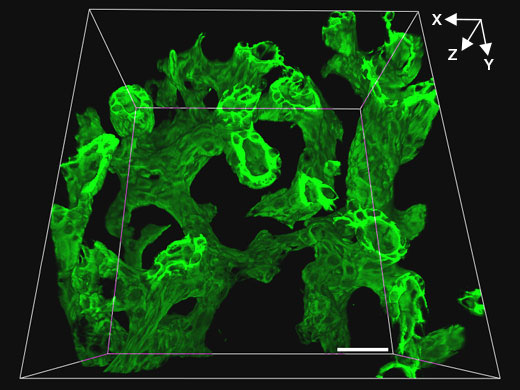 Control mouse | 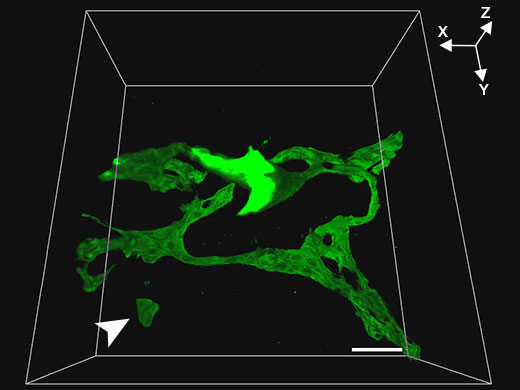 Klf5-LKO mouse |
Related VideosMovie: The biliary tree structure of the control mouse | To obtain higher resolution three-dimensional images, the FV3000 microscope and an Olympus 30x silicone oil immersion objective were used to obtain consecutive tomographic images (Z axial interval of 1 µm) of biliary tissue (green, biliary epithelial cell marker CK19) in 200 µm thick liver tissue cleared using SeeDB. This combination enabled high-resolution observation of the biliary trees of control and Klf5-LKO mice while maintaining a wide field-of-view. In the Klf5-LKO mouse, researchers observed CK19+ cell clusters (white arrow) that were spatially separated from the biliary tree. |
Comment from Dr. Okada:
Three-Dimensional Visualization and Fine Structural Analysis of Complex Biliary Structures
Dr. Hajime Okada | In this experiment, we analyzed 3D structures of biliary trees related to liver remodeling following a diet-induced liver injury. The biliary structure in cleared liver tissues in Klf5-LKO mice and control mice were compared using an FV3000 microscope. The microscope’s highly sensitive detector enabled the acquisition of wide field-of-view images with high resolution and brightness as well as fast observations with fewer images and averaging, resulting in many images that could be used for quantitative analysis. Furthermore, the Olympus silicone oil immersion objective enabled imaging at depth with high resolution, enabling us to find CK19+ cell clusters that were spatially separated from the biliary tree. These results suggest that biliary structure in tissue remodeling under various conditions of liver injury may be regulated by some molecular mechanisms. |
Comment from Dr. Itoh:
Three-Dimensional Imaging of Cleared Livers2)
Dr. Tohru Itoh | Until now, medical and biochemical research on the liver has mostly relied on conventional 2D observation methods using tissue sections instead of whole intact organs. Using these methods, however, it is challenging to detect and understand the “true colors” of the liver under physiological conditions and various conditions of liver diseases. Our research group developed new visualization technology for staining and clearing liver tissues, with which the 3D observation of the biliary structure in intact mouse livers was successfully realized for the first time. This combination of clearing and staining technology enabled us to detect dynamic biliary structural changes (biliary remodeling) and conduct research on its regulatory mechanisms and physiological functions. In those experiments presented above, we successfully established an experimental system for effective visualization of a 3D biliary structure using the SeeDB clearing reagent and a confocal microscope. By using an FV3000 microscope, some of the molecular mechanisms for biliary remodeling were clarified. With the techniques presented here, a confocal microscope is useful for 3D analysis of dynamic changes of various tissues and cells in the liver, including the biliary structure. We expect that our new visualization technology can contribute to the development of diagnostics and therapies for liver diseases and regenerative medicine through a better understanding of the process of development and regeneration from the viewpoint of tissue remodeling and inter-cellular interactions. 2) References: K. Kamimoto, K. Kaneko, CY. Kok, H. Okada, A. Miyajima, and T. Itoh, "Heterogeneity and stochastic growth regulation of biliary epithelial cells dictate dynamic epithelial tissue remodeling," Elife, 2016 Jul. 19;5, pii: e15034, doi: 10.7554/eLife.15034. K. Kaneko, K. Kamimoto, A. Miyajima, and T. Itoh, "Adaptive remodeling of the biliary architecture underlies liver homeostasis," Hepatology, 2015 Jun.;61(6):2056-66, doi: 10.1002/hep.27685, Epub 2015 Apr. 22. |
How the FV3000 Confocal Microscope Facilitated Our Experiment
Fully Spectral System Provides High Sensitivity
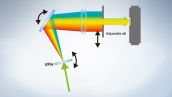
The FV3000 series employs Olympus' TruSpectral detection technology that diffracts light via transmission through a volume phase hologram unit. This technology enables much higher light throughput when compared to conventional spectral detection units with reflection-type gratings, and minimizes the laser power needed for deep tissue observation.
Silicone Immersion Objectives for Live-Cell Imaging Deliver High-Resolution Observation at Depth

The refractive index of silicone oil (ne≈1.40) is close to that of living tissue (ne≈1.38), enabling high-resolution observations deep inside living tissue with minimal spherical aberration caused by refractive index mismatch. Silicone oil also does not dry out or harden, so there is no need to refill the oil, making it ideal for extended time-lapse observations.
Acknowledgments
This application note was prepared with the help of the following researchers:
Division of Mammalian Development, Genetic Strains Research Center, National Institute of Genetics, Dr. Hajime Okada
Laboratory of Stem Cell Therapy, Institute for Quantitative Biosciences, The University of Tokyo, Project Associate Professor, Dr. Tohru Itoh
Products Related to This Application
was successfully added to your bookmarks
Maximum Compare Limit of 5 Items
Please adjust your selection to be no more than 5 items to compare at once
Not Available in Your Country
Sorry, this page is not
available in your country.





