Validation of Multipoint Observations Using an Incubation Monitoring System
Introduction
During the daily cell culture process, it takes a lot of time and effort for operators to properly maintain and control the culture conditions. To properly manage cells in a conventional cell culture workflow, operators must remove cells from the incubator and check their condition over time by observing and calculating cell counts and confluency under a microscope.
In addition, cell quality must be maintained at a constant level during the preparation stage to improve the consistency and reproducibility of experiments. However, differences in operational quality can occur depending on the skill level of the operator and other factors. Since cell culture is the starting point of life science and medical research, cell quality has a significant impact on subsequent experiments. Consequently, cell quality must be properly maintained and managed based on certain culture conditions.
Our CM30 incubation monitoring system simplifies this process by automatically and remotely monitoring cell culture conditions from inside the incubator, 24 hours a day, 365 days a year. It can also regularly visualize cell conditions based on AI-based analysis results. With continuous and standardized measurements, the CM30 system enables operators to quantify cell status efficiently and reproducibly without relying on their own
intuition.
Estimating the Cell State of the Entire Vessel from Multipoint Observations
To maintain cell quality, it is important for operators to observe and properly manage cell conditions daily. In the past, operators spent a great deal of time and effort to observe the entire container using a microscope to understand the cell condition and the appropriate timing for passaging and collection. At this time, even a somewhat skilled operator needs to check the observation field inside the container and the entire culture vessel because the seeding state of cells may be uneven depending on the cell type handled, the vessel, and the method of operation.
For this reason, the CM30 system can automatically image the entire vessel once a day, and then repeatedly image any specified observation point, enabling the camera to continue imaging the desired observation position while periodically acquiring information on the entire culture vessel.
However, collecting information on the entire container at each time point in the culture process involves various risks in terms of both hardware and software. For example, the handling of data is complicated due to the large amount of acquisition time and data volume. Further, the temperature inside the incubator rises due to the prolonged operation of the monitoring system, and the effects of these environmental changes on the cells must be considered.
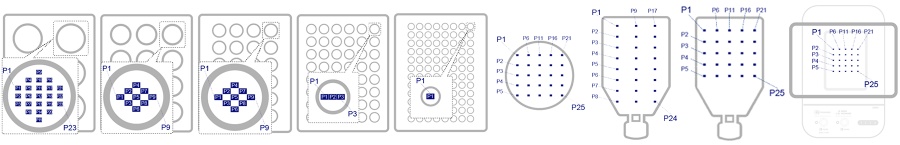
To simultaneously achieve data reliability and avoid these risks, the CM30 system counts the number of cells at multiple time points (Figure 1) defined according to the vessel size at each time point, then estimates the culture status of the entire vessel from the average of the analysis results. This enables the CM30 system to efficiently acquire quantitative data with little error from the actual culture conditions.
In this paper, we present the results of quantitative data acquisition, comparison, and verification* with the CM30 system to confirm the validity of estimating the cell state of an entire vessel from multiple points.
*This paper presents the results of an internal verification by Evident.
Figure 1. Number of multipoint observation points in different cell culture vessels.
Comparative Verification of Cell Count Estimates
The following cell types and culture vessels were used for validation to compare the results of the analysis on cell counts.
Cell type, culture vessel (number of multipoint observation points)
- A549, 12-well plate(9 points per well)
- NHEK, T75 flask(25 points)
Specifically, the same analytical parameters were applied to the entire vessel and to each of the multiple observation points defined for each vessel in CM30 system at the same time point, and the cell count results were compared. To compare the results across the entire cell proliferation process, we also validated the results for each of the three cell densities: low, middle, and high.
The validity of the CM30 system's multipoint observation was verified by comparing the cell count for the entire vessel, which was estimated from the average of the cell counts analyzed at multiple points defined for each vessel, and the cell count estimated by observing the entire vessel as the true value for the above three cell densities.
To verify the degree of variation in cell counts when estimating the entire container from only one observation point, we compared the cell counts of the entire container estimated from the maximum and minimum cell counts at the one observation point obtained by the above overall observation and the actual cell counts (true values) for the entire container.
Relevance of Multipoint Observations Using an Incubation Monitoring System
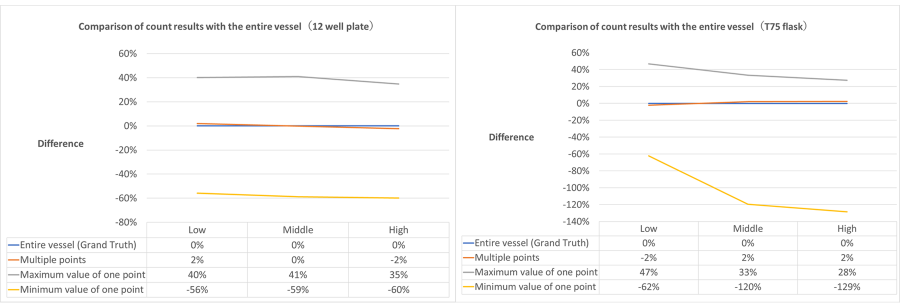
Figure 2 below shows a comparison of count estimates (true values) for the entire container and count estimates obtained from multipoint observations in a 12-well plate and T75 flask.
Figure 2. Error in cell count estimates between whole vessel and multipoint observation.
Typically, cell count and confluency analysis is performed for quality inspection during the cell preparation process with an accuracy of ±10% of the true value. In this validation, for both the 12-well plate and the T75 flask, the error between the whole container and the estimate based on multipoint observation was within ±2% throughout the entire growth process.
Therefore, we found that the estimates obtained from multipoint observations of each vessel provided sufficiently reliable data without the need to observe the entire vessel. Even under severe conditions such as a 12-well plate with a small number of observation points per vessel area, the error in the estimated values for the entire vessel was within ±2% for the entire growth process. Therefore, the number of multipoints of the CM30 system was considered sufficient for estimating the cellular condition of the entire vessel.
In addition to the validation of multipoint observation, we checked the same method when we counted the number of cells at only one observation point in a container and estimated the cell status of the entire container based on the results (Figure 2). We found that a counting error of up to 60% occurred in a 12-well plate and a counting error of up to 129% occurred in a T75 flask.
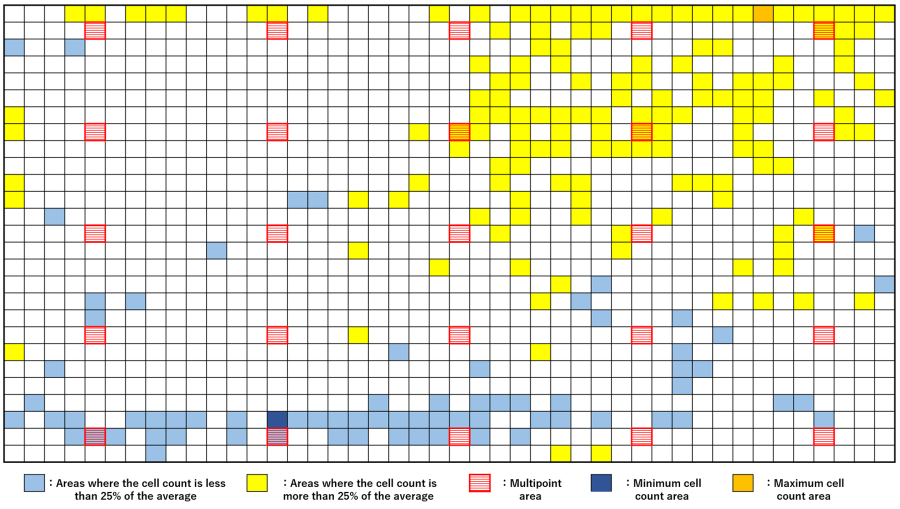
To verify whether this result was due to uneven cell seeding, we used a heat map of the degree of variation in cell counts across vessels in the T75 flask (Figure 3). Yellow indicates cell areas with more than 25% more cells than the overall average cell count, and light blue indicates areas with less than 25% of cells. The red line indicates the CM30 multipoint observation area (T75 flask), orange indicates the area where the most cells were detected, and blue indicates the area where the least cells were detected.
The results of the verification show that even a somewhat skilled operator who routinely performs cell culture can experience this kind of seeding variation in parts. It also shows that the risk of overall estimation is very high if a single observation point is covered by an area that deviates from the average cell count of the entire vessel.
Figure 3. Heat map showing the degree of variation in cell counts in the T75 flask.
As seeding irregularities occur at a certain rate as shown above, estimating the cell state of the entire vessel from a single observation point carries a high risk of obtaining results that differ significantly from the actual cell count—even if there are no major issues in the operator's cell culture operation. This demonstrates that it is difficult to obtain quantitative data from a single observation point with high reproducibility. For example, when using a monitoring device that
is limited to a single observation point, it is necessary to observe multiple points by moving the container position each time to quantify the cell state of the entire container.
Conclusion
Our comparative validation of the CM30 system shows that it is possible to estimate quantitative data close to the actual cell count of the entire container from the average of cell counts from multiple points without observing the entire container. This demonstrates the validity of estimating the entire container from multiple points. In addition, considering that a certain amount of seeding irregularities occur even when the operator is somewhat skilled, estimating the cell count of the entire vessel from a single observation point has a high risk of obtaining data that is significantly different from the actual cell count.
The results of this study suggest that quantitative data obtained by multipoint observation on the CM30 system is reliable for cell quality control. They also suggest that even when simply checking the cell condition, it is important to consider a certain degree of seeding irregularity and perform multipoint observation.
In addition to the above analysis methods, the CM30 system has been updated to improve cell count, confluency, and other analysis functions. The accuracy of the AI-based analysis has also been enhanced with the addition of more detailed settings for each analysis parameter. As the CM30 system focuses on efficiency and reproducibility by quantitatively monitoring culture conditions while minimizing human variability, it is an optimal solution for cell culture processes with a large number of
daily tasks.
Author
Masatoshi Dehari
Life Science Research Solutions, Global Marketing
Evident
Products Related to This Application
was successfully added to your bookmarks
Maximum Compare Limit of 5 Items
Please adjust your selection to be no more than 5 items to compare at once
Not Available in Your Country
Sorry, this page is not
available in your country.