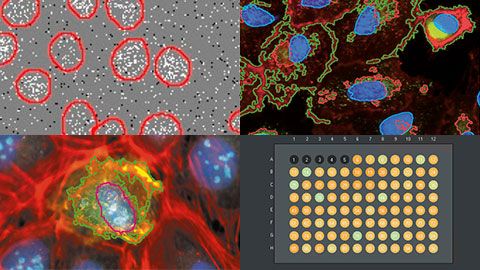
In this webinar, Manoel and Shohei, our deep learning and high-content screening experts, will introduce the scanR system’s assay builder. The scanR system is Olympus’ dedicated high-content screening platform with a unique sample navigation approach and analysis inspired by flow cytometry.
FAQWebinar FAQs | High-Content Screening: Customized Analysis Made EasySome cells present with two different colors (e.g., green and blue), suggesting two different cell-cycle stages. Is this an indication that the model is not performing correctly?The system can differentiate between multiple colors/channels, so when two different colors are present, the software identifies background from foreground (where the foreground is the cell layer). However, because some cell-cycle stages can be identified with morphological features, we strongly suggest using a cell-stage tag to mark different cell stages. This will be integral to ensure that training (deep learning) is performed with all cell-cycle stages that you want to differentiate. Nevertheless, since cells are evaluated per pixel and not “per cell,” there are scenarios where the system will be able to automatically discern cell-cycle stages, even if you did not specifically train with that stage. How does the system perform when analyzing spheroid cultures or other cultures?The scanR system is designed to efficiently work with spheroid cultures or other types of culture as well. There are some workflow details that we have observed to work well with this application. First, we recommend you apply 3D deconvolution to images collected, and then carry out manual annotation. An example workflow would involve using 10–20 images and manually segmenting 10 cells. Can you image the borders of the wells in 384-well plates?Yes, but there are limitations. The ability to image well borders is dependent on the combination of the type of well plate and the objective used. Well plates come in a variety of forms, including skirted, semi-skirted, and non-skirted. Therefore, the important information when imaging well borders is the difference in height between the bottom of the plate and the bottom of the skirt. As such, there comes a point where the vertical distance of the bottom of the plate to the bottom of the skirt prevents a high magnification objective from imaging, because the objective may collide with the plate. In this case, we strongly suggest a long-working distance objective. Specifically, Olympus offers a great range of long-working distance objectives. How is focusing done in the scanR system?The scanR system uses a combination of hardware and software autofocus. The hardware autofocus detects the bottom surface of the cell, and the software autofocus finds the sample portion. This is a very robust process, so you can basically walk away from the system once you’ve started the experiment. Can you use the super-resolution spinning disk system for high-content screening?Yes, you can use the scanR software to control the Olympus IXplore SpinSR microscope system. It is possible to achieve 120 nm resolution with this system and then import acquired images into cellSens software for further processing and even better results. How well does the segmentation and deep learning perform on tissue slices?In our experience, segmentation works well on tissue slices, especially when using the spinning disk system. However, it is possible to acquire good images without the spinning disk system by just applying 3D deconvolution. If tissues are particularly difficult to work with, then deep learning with manual annotation has been shown to work well. Manual annotation provides the deep-learning algorithm a robust training set from which to learn and better predict segments, based on user identification. |