Comparing Human iPS Cell Lines using the CM20 Incubation Monitoring System Part 2: Variations in the Efficiency of Liver Bud Organoid Differentiation among iPS Cell Lines
Introduction
Induced pluripotent stem (iPS) cells are widely used not only for basic research such as developmental biology focusing on cell differentiation and organ formation, but for translational research including drug discovery and the development of diagnostic tools. The technology of constructing human iPS cell-derived mini-tissues/organs—called organoids—has rapidly progressed to the point that three-dimensional tissues can be utilized as an avatar of human organs. To date, many iPS cell lines established both from healthy subjects and patients worldwide can be utilized for organoid studies, which aim to clarify the genomic background of diseases and the predisposition to diseases for each patient by evaluating the functional and individual differences of the generated organoids. However, the process of generating organoids of the target organs generally requires more than one month and is expensive. The stability and reproducibility of organoid generation still remains a major challenge due to the variability in differentiation ability and proliferation rate among the iPSC cell lines used. This is one of the barriers to the development of organoid research based on the comparison of multiple samples derived from different iPS cells.
To solve this issue, we quantitatively monitored iPS cells over a long period of time to better dissect differences in the undifferentiated state and their relationship to cell line-dependent variation in organoid differentiation.
Among human iPS cell lines derived from 12 donors, certain lines often failed to differentiate into liver bud organoids in spite of the same differentiation protocol applied across all cell lines tested. In this application note, we conducted an experiment to better understand the relationship of iPS cell lines between the properties at undifferentiated states before differentiation induction and differentiation potential into liver bud organoids.
Using iPS cell monitoring data from the CM20 system to evaluate organoid differentiation
Olympus’ CM20 incubation monitoring system enabled us to quantitatively measure the culture status and acquire measurement data that could easily be compared to past results. The iPS cell colony number and density data can be easily exported to a CSV file for more detailed analysis.
In this study, we monitored the growth of human iPS cell lines from 12 donors during their maintenance, and then exported time-dependent colony number and density data for each cell line. We then applied the differentiation protocol that we have recently developed (*1) to obtain liver bud organoids. After about one month, we counted the number of generated organoids and measured the level of albumin secretion as a function of liver bud organoid. We also compared the iPS cell growth data obtained before differentiation with the data of their corresponding liver bud organoids for each cell line.
Culture Protocol and Data Analysis
Twelve human iPS cell lines were maintained in a 6-well plate under feeder-free conditions. Approximately one week after passage, the cells were dissociated into single cells with Accutase and then seeded on a new plate. They were stimulated with Activin A and BMP4 to differentiate from undifferentiated states into definitive endoderms. We then treated them with FGF4 and Wnt agonist to obtain posterior foregut spheroids. To shift into a three-dimensional culture, we embedded these spheroids in Matrigel, a basement membrane matrix, followed by pulse of retinoic acid (RA) during the early stage of organoid formation. The cells were further cultured in a hepatic differentiation medium for up to about one month, resulting in the generation of liver bud organoids (Figure 1A).
We next counted the number of organoids and measured the levels of albumin secretion with an ELISA of the collected medium samples. We integrated the undifferentiated iPS cell proliferation data that was obtained using the CM20 monitor and analyzed the correlation between the characteristics during the maintenance culture and the organoid differentiation efficiency of each iPS cell line (Figure 1B).
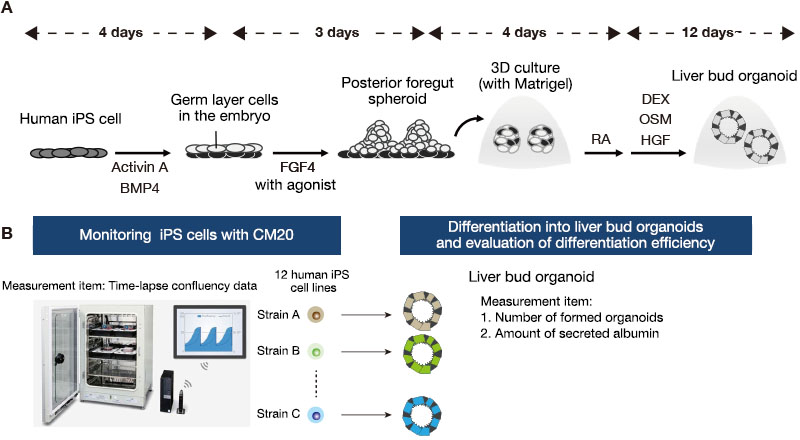
Figure 1. Monitoring human iPS cells during maintenance culture and evaluating liver bud organoid differentiation efficiency.
(A) The culture protocol used to differentiate human iPS cells into liver bud organoids.
(B) Summary of comparative analysis of characteristics during iPS cell maintenance and liver bud organoid differentiation efficiency across 12 iPS cell lines derived from different donors.
Throughout multiple attempts to induce 12 human iPS cell lines to differentiate into liver bud organoids, we were able to obtain organoids from most of the cell lines. However, iPS cell lines I and J often failed to differentiate (Figure 2A). Although the albumin secretion (the amount of secretion per organoid) varied depending on the differential induction batch, the liver bud organoids derived from I and J iPS cell lines—which have extremely low organoid formation efficiency—also showed low levels of albumin secretion (Figure 2B). Among the cell lines with high organoid formation efficiency, we noticed some iPS cell lines, such as line C, showing relatively low albumin secretion. To further validate the differentiation, we confirmed that liver bud organoids derived from iPS cell lines other than I and J expressed hepatocyte-specific markers (data not shown). These data suggested that among the 12 cell lines tested, I and J iPS cell lines have restricted capacity to differentiate into the liver lineage.
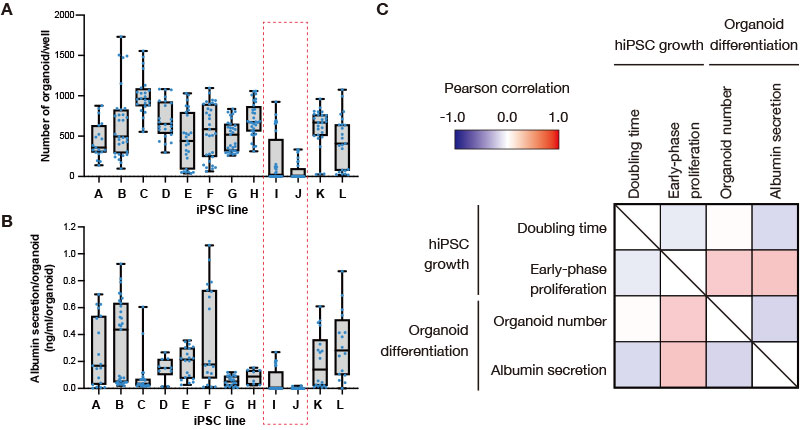
Figure 2. Correlation analysis of cell proliferation before differentiation induction and the efficiency of liver bud organoid differentiation across human iPS cell lines.
(A, B) The number of liver bud organoids formed during the multiple differentiation induction experiments (A) and the level of albumin secretion (B) were plotted in a box plot for each iPS cell line. Blue dots correspond to the number of wells measured during multiple independent experiments.
(C) The correlation map showing growth parameters of human iPS cells during maintenance (doubling time and early-phase proliferation) and liver bud organoid differentiation (organoid number and albumin secretion).
Next, to investigate whether the state of iPS cells before the differentiation induction step affects the liver bud organoid differentiation efficiency, we analyzed the correlation of the proliferation parameters acquired by the CM20 monitor, the number of formed organoids, and albumin secretion corresponding to each differentiation induction experiment. We calculated correlation coefficients between the iPS cell doubling time before differentiation induction (calculated using confluency data provided by the CM20 monitor), the growth efficiency at the initial stage of proliferation (calculated from a formula defined by Dr. Takebe), the number of generated liver bud organoids, and the levels of albumin secretion for each experiment. We accounted for batch-dependent variations by analyzing the dataset of each experiment (Figure 2C). While the number of generated liver bud organoids and the levels of albumin secretion showed almost no correlation with the iPS cell doubling time, they were positively correlated with the parameters of proliferation efficiency in the early stage of iPS cell proliferation. This indicates that although simple differences in doubling time between iPS cell lines cannot be used to determine whether the liver bud organoids generated are good or bad in their qualities, it is possible that some differences at the early stages of iPS cell proliferation observed between cell lines affect the efficiency of subsequent liver bud organoid differentiation.
Conclusion
The CM20 monitor’s quantitative data enabled us to look at information from past cell cultures up to and even after differentiation into organoids. The images and data are easy to export, enabling a variety of analyses that can lead to new insights during the culture process.
In this study, we found that the variable efficiencies of liver bud organoid differentiation across iPS cell lines may be significantly affected by the growth efficiency of the iPS cells during their maintenance, particularly in the early stage of growth. This suggests that to generate more stable and higher quality liver bud organoids, it is important to monitor the state of iPS cells immediately after passage to know whether they are ready for differentiation induction. Based on this correlation, it is also possible to predict when a cell line will have good or poor differentiation efficiency by monitoring the proliferative state of iPS cell lines.
Olympus will continue research using CM20 monitor to develop methods for efficient quality control and data management.
Comments from Dr. Takebe and Dr. Yoneyama
Dr. Takanori Takebe (left)
| While handling multiple iPS cell lines derived from different donors and evaluating the differences between donors using iPS cell-derived organoids, we frequently encounter the problem that differentiation efficiency varies among iPS cell lines. The advantages of the CM20 monitor, including stable monitoring of proliferation states and the ability to export the data, were very helpful for us to perform the correlation analysis between the proliferation of iPS cell lines and differentiation efficiency of the generated organoids in 12 cell lines tested simultaneously. |
References:
*1) Koido M et al., Nature Medicine 2020 (PMID: 32895570), Takebe T et al., Nature 2013 (PMID: 23823721), Takebe T et al., Cell reports 2017 (PMID: 29212014)
Products Related to This Application
was successfully added to your bookmarks
Maximum Compare Limit of 5 Items
Please adjust your selection to be no more than 5 items to compare at once
Not Available in Your Country
Sorry, this page is not
available in your country.
