Comparing Human iPS Cell Lines using the CM20 Incubation Monitoring System Part 1: Observing Differentiation into iPS Cell-Derived Liver Bud Organoids
Introduction
Induced pluripotent stem (iPS) cells are widely used for basic research on embryology, such as cell differentiation and organ formation, to drug discovery research and developing diagnostic methods for diseases. To date, many iPS cell lines have been established globally, but their properties—such as differentiation potential and growth speed—differ among the strains. While these differences are known, they have never been quantified using cells grown under the same environmental conditions. This makes it difficult to determine whether these variations are due to differences in environment and procedures between labs or whether they are inherent in the cell lines themselves. Even if there are variations in the properties of differentiated cells derived from iPS cells, they are not discovered until the culture is completed, so cells that have been induced to differentiate may have abnormalities. Against this background, it's important to have a tool that enables long-term iPS cell monitoring and efficient quality control and data management of the process up to the differentiation induction experiment.
Quantitative Cell Culture Data Analysis Using the CM20 Incubation Monitoring System
With the quantitative measurements provided by the Olympus CM20 incubation monitoring system, we analyzed control points for variations in organoid creation by iPS cells and established a method for analyzing the disease genome background based on multi-specimen comparison. In this study, 12 human iPS cell strains established from different donors were cultured using the same process, and data on the growth process of each cell line were obtained using the Olympus CM20 incubation monitoring system.
Culture protocol
Maintenance of the human iPS cell cultures was performed in a 6-well plate under feeder-free conditions using StemFit AK02N medium (Ajinomoto Co., Inc.) and iMarix-511 (Matrixome Inc.) plate-coating matrix. They were subcultured using unicellularizing with a detachable Accutase solution with seeding at 1.0–2.0 × 104 cells / well, and cultured in StemFit AK02N medium supplemented with Y-27632 for 24 hours. After, the human iPS cells were proliferated by exchanging with StemFit AK02N medium with no Y-27632 and then cultured for about 1 week while exchanging the medium once every two days (Fig. 1 ).
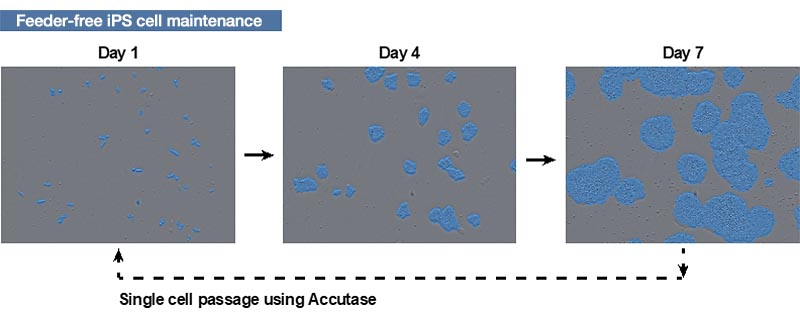
Figure 1. How human iPS cells are cultured
Images of human iPS cells acquired by the CM20 monitor on days 1, 4, and 7 after passage. The blue area was identified by the monitor’s automatic colony recognition function.
The blue area was identified by the monitor’s automatic colony recognition function.
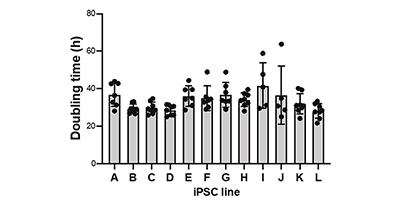
For the 12 human iPS cell strains, the growth process during 5 to 8 passages was continuously monitored with the CM20 system (Fig. 2 A, B). These iPS cell lines underwent medium exchange and subculture at the same time. We found that each iPS cell proliferated in a doubling time of about 30 hours, and the proliferation speed was slightly different between iPS cell lines. We also found that among the human iPS cell lines, there are iPS cell lines (such as B strain and C strain) whose doubling time is almost constant between passages. This confirms that culture processes, such as medium replacement and subculture, were performed correctly (Fig. 2 C).
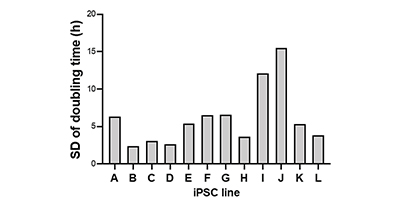
On the other hand, some iPS cell lines, such as strain I and strain J, showed large variations in doubling time for each passage (Fig. 2D). These results demonstrate that the 12 iPS cell lines have different growth properties.
A
| B
|
C
| D
|
Figure 2. Quantitative proliferation status monitoring during human iPS cell culture maintenance
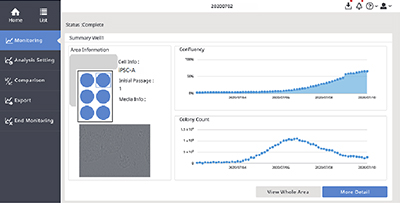
(A) CM20 monitor software interface.
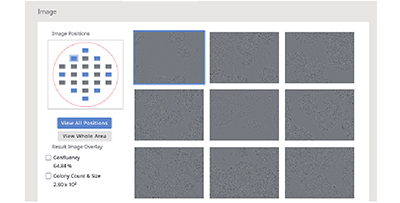
(B) Single-well observation using the CM20 monitor. Images of 9 fields of view in the wells are acquired every 3 hours, and all well scans are automatically performed every day.
(C) The doubling time for each passage performed multiple times is plotted for each iPS cell line. The mean ± standard deviation (SD) is shown in the bar graph.
(D) The variation in doubling time for each iPS cell line are shown in SD.
Quantitative analysis advantages
Because the CM20 monitor collects quantitative data on the cell growth process, data from different cell lines can be measured and compared using the same standards. The monitor makes it easy to collect data from multiple strains maintained by a single researcher under the same process as well as from and multiple researchers. This makes it easy to check if the process was done correctly by comparing cell images and growth.
In the past, when cell quality varied, it was difficult to determine whether it was caused by differences in the properties of individual cell lines, or an error in the culture process is to be the causes of variation in the cell culture process when the quality varied. In particular, many studies using iPS cells require a lot of work to obtain the desired differentiated cells. It was difficult to measure how changes that can occur during iPS culture maintenance affect differentiated cells’ evaluation results.
Olympus will continue research using CM20 with the aim of developing methods for efficient quality control and data management.
Comment from Dr. Takebe and Dr. Yoneyama
Dr. Takanori Takebe (left) | With the CM20 monitor, which can be permanently installed in the incubator, 6 to 12 iPS cell strains can be constantly monitored simply by placing the culture vessel or plate on the monitor. The growth data is captured and recorded without having to remove the cells from the incubator. It was useful for acquiring data on the differences between the cell lines. In addition, it takes a lot of effort to manually take photos of multiple stocks over time, but one of the advantages of the CM20 monitor is that it automatically stores and organizes the photos. |
Products Related to This Application
was successfully added to your bookmarks
Maximum Compare Limit of 5 Items
Please adjust your selection to be no more than 5 items to compare at once
Not Available in Your Country
Sorry, this page is not
available in your country.