Perform Accurate and Efficient Microscopy Image Analysis Using TruAI based on Deep Learning
Introduction
Experiments often require data from microscope images. For accurate image analysis, segmentation is important to extract the analysis target area from the image. A common segmentation method is to apply thresholds to the image intensity values or color.
While effective, this method can be time-consuming and affect the sample condition. Next-generation image analysis methods like our cellSens imaging software with deep-learning-based TruAI help reduce the risks of sample damage while achieving high efficiency and accuracy.
Application Examples Using TruAI
1) Label-Free Nucleus Detection and Segmentation
To count the number of cells, localize nuclei in cells and tissues, and evaluate the cell area, researchers commonly use fluorescent staining of nuclei to segment based on the fluorescence intensity information.
In contrast, TruAI can perform nucleus segmentation using only brightfield images. It works by training a neural network using the nucleus segmentation results from brightfield and fluorescence images.
This self-learning microscopy approach eliminates the need to fluorescently stain the nucleus once the neural network is created. Other benefits include:
- Minimize time spent on nuclear labeling
- Exclude effects on cells due to labeling
- Prevent phototoxicity and fading
- Acquire extra sample information by adding another channel
 Label free nucleus detection by TruAI Figure 1 |
Figure 2 |
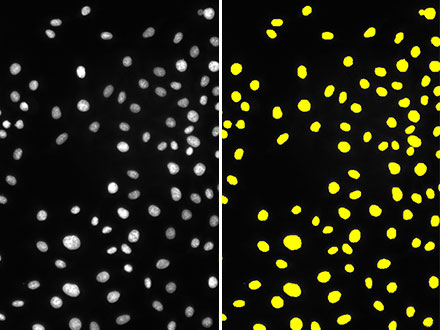
Figure 1: While the brightfield image (left) has minimal contrast due to unstained cells, the TruAI detects the nuclei with high accuracy (right).
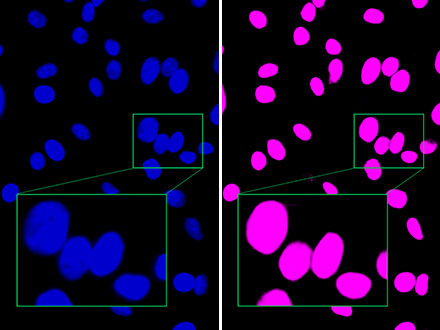
Figure 2: Compared to the fluorescence image (left), Olympus’ TruAI clearly distinguishes close nuclei from one another (right), indicating that detection is possible with high accuracy.
2) Quantitative Analysis of Fluorescently Labeled Cells with Ultra-Low Light Exposure
Fluorescent labels are an invaluable tool in modern microscopy-based cell studies. However, the high exposure to excitation light can lead to photodamage or phototoxicity and have an observable impact on cell viability. Even if no direct effect is observed, strong light exposure can influence the cells’ natural behavior, leading to undesired effects.
In long-term live cell experiments, minimal light exposure during fluorescence observation is ideal. From a technical standpoint, ultra-low light exposure means analyzing images with very low signal levels and, consequently, low signal-to-noise ratios. Our TruAI enables you to analyze low-signal images with both robustness and precision.
Figure 3 |
Figure 4 |
Figure 5 |
Figure 3: The result of detecting nuclei (right) from a fluorescence image (left) with sufficient luminance using a conventional method that applies a luminance threshold.
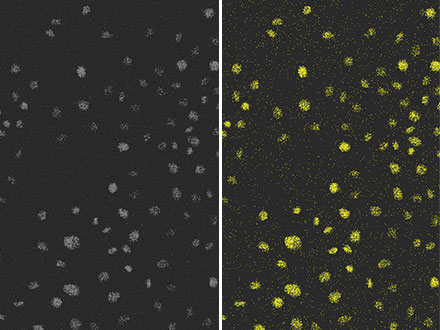
Figure 4: The result of detecting nuclei (right) with the same conventional method as Figure 3, from fluorescence images (left) with extremely poor SNR due to weak excitation light. You can see that the detection accuracy is low.
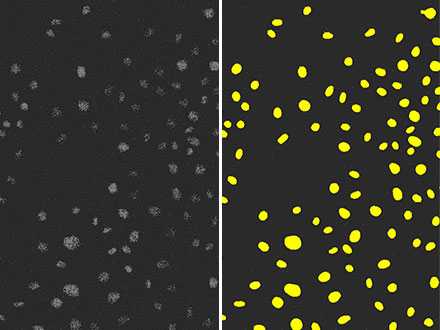
Figure 5: The result of detecting nuclei (right) using TruAI from a fluorescence image (left) with extremely poor SNR due to weak excitation light. You can see that the accuracy is as high as Figure 3 and performed with much higher accuracy than Figure 4.
3) Segmentation Based on Morphological Features
If you want to segment an image based on its morphological features, it is very difficult to achieve high-precision segmentation with the conventional approach of applying thresholds to intensity values and color. Therefore, it was necessary to manually count and measure each time.
In contrast, TruAI enables highly efficient and accurate segmentation based on morphological features. After the neural network learns the segmentation results from hand-labeled images, it can apply the same methodology to additional data sets. For instance, neural networks trained from hand-labeled images can count mitotic cells, as depicted in the images below.
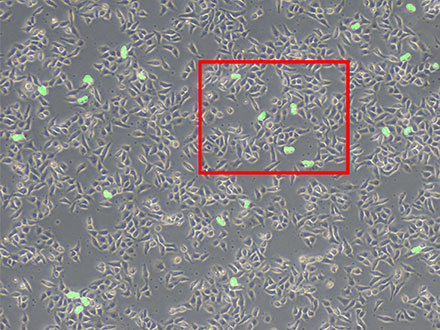
Figure 6 |  A high-magnification image (left) of the framed area in Figure 6 Figure 7 |
Figure 6: Prediction of mitotic cells using TruAI (green).
Figure 7: While you can see many cells, only the dividing cells are detected (right).
4) Tissue Specimen Segmentation
TruAI can also be used to segment tissue specimens. For example, kidney glomeruli are difficult to discriminate using conventional methods but can be segmented using TruAI.
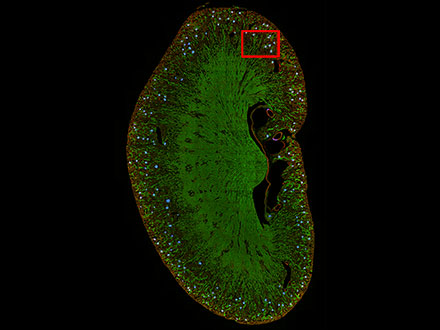
Figure 8 |  A high-magnification image (left) of the framed area in Figure 8 Figure 9 |
Figure 8: Prediction of glomeruli positions on a mouse kidney section using TruAI (blue).
Figure 9: TruAI captures and detects the glomeruli features (right).
Conclusion
Conventional segmentation methods can be difficult and cause damage to samples. Our cellSens imaging software with deep learning enables accurate and efficient segmentation in conditions that cause minimal damage to cells, such as label-free imaging or ultra-low light exposure. The software also makes it easier to perform segmentation of tissue specimens based on their morphological features.
Products Related to This Application
was successfully added to your bookmarks
Maximum Compare Limit of 5 Items
Please adjust your selection to be no more than 5 items to compare at once
Not Available in Your Country
Sorry, this page is not
available in your country.