Advanced Live-Cell Analysis Using AI-Driven High-Content Screening Systems
Introduction
Artificial intelligence (AI) capabilities in modern high-content screening systems are opening doors in live-cell analysis.
This application note explores how AI-powered systems like the Olympus scanR high-content screening (HCS) station are overcoming common fluorescence microscopy challenges and helping to enable advanced analyses of living cells that previously seemed impossible. Learn how these systems can provide greater precision, ease of use, data quality, and cell viability.
Observing Biological Samples without Markers
In the observation and analysis of biological characteristics and processes, the labeling of cells with color markers, especially fluorescent markers, is invaluable today. Historically, however, the observation of biological samples without markers was the original analysis method used in microscopy. This method has become important again due to revolutionary advances in image analysis through machine learning. Deep-learning-based approaches of AI can provide better access to information contained in transmitted light images, making the fluorescent markers that are currently used for staining cells or cell components unnecessary.
An important reason for the low prevalence of label-free assays is that there were no methods available that could overcome the obstacles in transmitted-light analysis (for example, low contrast, especially in brightfield imaging) or—unlike fluorescence observation—the negative effects of dust and other contamination in the beam path on the image quality. When imaging living cells in microwell plates, it can be particularly difficult to generate transmitted-light images of sufficient quality for analysis due to various interferences, such as meniscus formation on the buffer medium. Using microwell plates has typically had many limitations and difficulties. For example, phase contrast observation is often impossible, brightfield images contain shading around the edges of the wells, and particles in suspension can cause an increase in background signal.
Deep-Learning Technology Opens Doors for Label-Free Analysis
Brightfield transmitted-light imaging is a straightforward approach for label-free analysis applications; however, image analysis and segmentation has long remained an unsolved problem. To solve this challenge, Olympus integrated image analysis based on a dedicated convolutional neural network (CNN) (Figure 1) into the scanR high-content screening software (Figure 2).
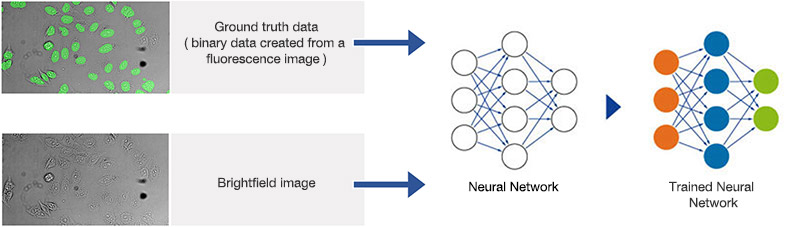
Figure 1A. Schematic illustrating the neural network training process.
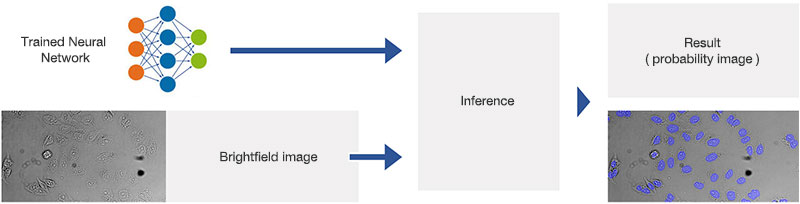
Figure 2A.Schematic illustrating the application (interference) of the trained neural network.
Neural networks of this kind are extremely adaptable to different and difficult image analysis tasks, making them an optimal choice for complex analysis of transmitted-light brightfield images of labelfree samples. In a training phase, the neural network automatically learns how to determine the desired parameters—for example, the position and contours of cells or cell compartments—a process that is referred to as the segmentation of objects of interest.
During the training phase, pairs of sample images and ground truth data (true basic data, which is, in this case, object masks with annotated objects of interest), are fed into the neural network. Once the network is trained, it can be applied to new images and predict the object masks with high precision. In conventional machine learning, the annotations (e.g., cell boundaries) are provided by human experts.
This can be a tedious and time-consuming step, because neural networks require large amounts of training data to fully exploit their potential. To resolve this issue, Olympus uses a concept called self-learning microscopy.
Figure 2.The Olympus scanR high-content screening station combines the modularity, flexibility, and robustness of a microscope-based design with the automation, speed, throughput, reliability, and reproducibility required for screening applications.
Understanding Self-Learning Microscopy
Self-learning microscopy means that the microscope automatically generates the ground truth data required for the training of the neural network by acquiring reference images during the training phase. For example, to teach the neural network to reliably detect and segment cell nuclei in brightfield images under difficult conditions, the nuclei can be marked with a fluorescence marker. The microscope then automatically acquires a large number of image pairs (brightfield and fluorescence).
These pairs are then used to train the neural network to reliably analyze new images. Since this approach of generating ground truth data requires hardly any human intervention and is scale independent, very large quantities of training image pairs can be acquired in less time. This training phase enables the neural network to learn how to deal with variations and distortions, which leads to a learned neural model that is particularly robust in meeting these challenges.
The Advantages of Using Artificial Intelligence for HCS Applications
For HCS applications, AI capabilities can be used to enable label-free quantification of living cells. The scanR HCS station’s software reliably determines the nuclei position in microwell plates using brightfield transmitted-light imaging—with an accuracy that is similar to that of fluorescence imaging (Figure 3).
Figure 3.. AI analysis of brightfield images. Comparison of the AI results (left) with a fluorescence staining (middle) superimposed on the brightfield image that the AI used for analysis (right).
The quantification of living cells using the brightfield method instead of the fluorescence method has many important advantages. First, it minimizes the exposure time and avoids the use of genetic modifications or nuclei labeling. This approach also leaves fluorescence channels available for other markers. Phototoxicity is reduced, leading to easier and faster image acquisition and improved cell viability (Figure 4).
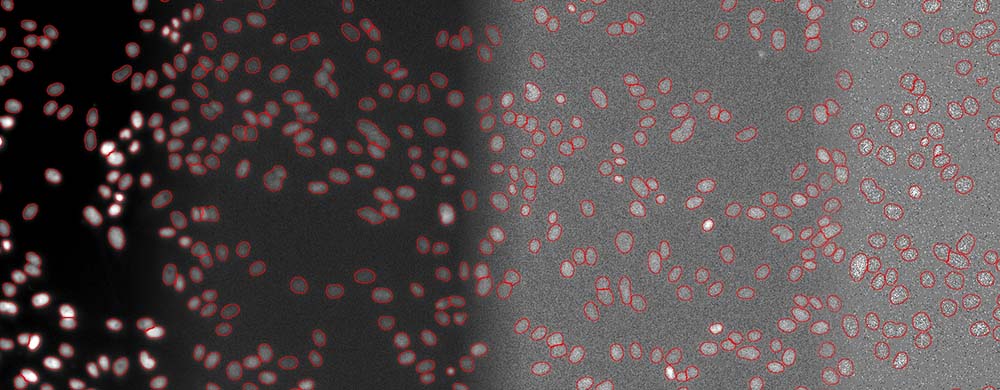
Fifure 4. Segmentation of cell nuclei (DAPI stain) for quantitative DNA post-analysis. Several exposure times (100%, 2%, 0.2%, 0.05%).
AI Analysis of Low-Light Samples
Due to the scanR system’s AI-based imaging software, reliable fluorescence analysis is also possible in low-light conditions. For example, the software recognizes DAPI-stained cells at only 0.2% of the optimal light intensity. It can even distinguish different stages of the cell cycle based on the signal intensity, enabling better information and optimized reproducibility.
This software capability offers important advantages for a wide range of life science applications. Analysis in low light and without labels is just one example of how automated detection using an AI-based approach can improve the accuracy and reproducibility of HCS. The self-learning microscopy approach combines these advantages with a user-friendly workflow that requires little human intervention during the training phase. As a result, users can exploit the full potential of HCS data and benefit from a quick and easy setup.
Conclusions
An AI-based approach offers significant advantages for many live-cell analysis workflows. In addition to increased precision, genetic modification and nuclei labeling are no longer required due to the brightfield imaging method. This not only saves time during sample preparation, but also reserves the use of the fluorescence channel for other markers. Further, the shorter exposure times in brightfield imaging and in analysis using low-light fluorescence conditions reduce phototoxicity and help save time.
Products Related to This Application
was successfully added to your bookmarks
Maximum Compare Limit of 5 Items
Please adjust your selection to be no more than 5 items to compare at once
Not Available in Your Country
Sorry, this page is not
available in your country.