DAPI illumination got you feeling burned? Fluorescence labeling is an invaluable tool for the observation and analysis of biological features, but it can come at the expense of cell health.
In the two-part video series below, we show you how to use cellSens software’s TruAI deep-learning technology to eliminate DAPI labeling from your experiments, reducing phototoxicity and removing a step from your sample preparation routine without sacrificing the data previously provided by DAPI channels.
Prepare Your Training Data
Related Videos
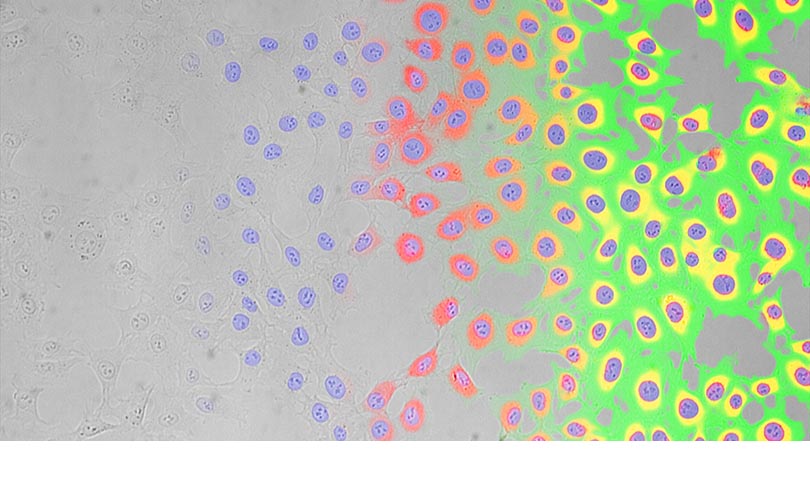
Before you start using TruAI deep-learning technology, you have to train the software’s neural network by providing it with ground truth data. The ground truth consists of a data set that provides the intended result in addition to the data needed to identify this result. For label-free nucleus identification, your ground truth data would be a series of images in which you’ve identified the nuclei.
In this video, we’ll show you the TruAI neural network is trained to identify nuclei using a dataset of phase contrast, brightfield, and DAPI images.
Using the Training Data to Identify Nuclei
Related Videos
Once the neural network is trained, it can be applied to a dataset with only phase contrast and brightfield. This is considered the inference phase, where a ground truth is no longer necessary to identify an object of interest.
No label-free approach can completely replace fluorescence, but eliminating DAPI from your experiments can:
- Reduce the sample preparation complexity
- Reduce phototoxicity
- Save fluorescence channels for other markers
- Enable faster imaging
- Improve the viability of living cells by avoiding stress from transfection or chemical markers
Related Content
Perform Accurate and Efficient Microscopy Image Analysis Using TruAI Technology with Deep Learning